The Electronic Laboratory Workbench (eLabBench) is an interactive laboratory workbench integrating wet-lab biology experimentation with digital bioinformatics analysis [1, 2, 3].
The eLabBench is a tabletop system supporting experimental research in the biology laboratory. The eLabBench allows biologists to organize their experiments around the notions of activities and resources, and seamlessly roam information between their office computer and the digital laboratory bench. At the bench, biologists can pull digital resources, annotate them, and interact with hybrid (tangible + digital) objects such as racks of test tubes.
The use of the eLabBench is shown in the video below (in Danish).
Technical Implementation
The main architecture and its components are shown in Figure 1: two client applications (the activityDock and the eLabBench) and a server infrastructure for activity roaming.
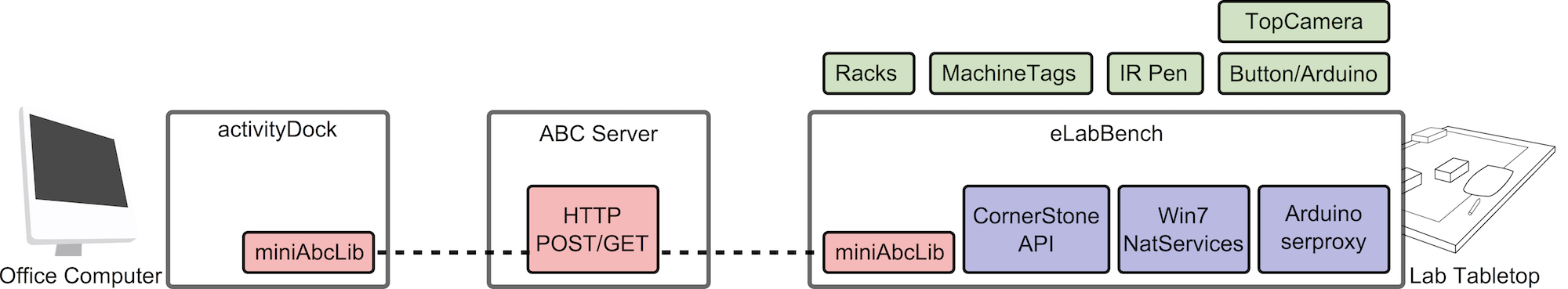
eLabBench
The eLabBench is a 46” Multitouch Cell2 encased in a specially designed laboratory bench and covered by a 6 mm thick glass plate. The main component of the Multitouch Cell is a 1920×1080 px LCD display, which can display very readable documents alongside (e.g. pdfs or web-pages). The bench-top measures 120×80 cm with an interactive screen surface of 105×60 cm. The bench has a margin of 10cm around the screen; this allows laboratory gas and water pipes to pass behind the bench, and provides minimal leg room on the front for users to sit. The bench is 93 cm high, which makes it possible to use in a standing position or sitting on a normal laboratory chair.
Additional components include a top mounted camera attached to a shelf standing above the bench and an infrared-LED pen. Users operate the camera through a big red button attached to the front of the bench. Initial testing showed that white color objects like paper, aprons (from the biologists elbows as it is common practice to rest the elbows on the bench while pipetting), boxes, etc, are highly IR-reflective and therefore are source of many false positive touches. To limit these false positive touch detections, we increased the threshold in the vision algorithm to only detect touches activated by an custom-made infrared pen. Users activate the light pen with a button.
The eLabBench runs Windows applications. On startup, it launches an almost full screen application (with only the windows toolbar left visible). It relies on the miniAbcLib for data management and the Cornerstone API for input (touch and marker tracking) (see Figure 1-right). The eLabBench integrates with two types of external applications: the laboratory’s digital notebook and native Windows 7 applications. We integrated with the Confluence wiki5 used in the laboratory we study. This allows biologists to update a protocol on the fly or add resources to the wiki while at the bench (for instance a picture from the top camera). Besides wiki integration, the eLabBench implements activity-based computing and can invoke and control external applications’ windows using a set of native Windows services.

The eLabBench implements object integration by augmenting two types of objects: test tube racks and machines tags. In the laboratory, test tubes racks come in a variety of shapes and sizes, therefore the eLabBench supports 5 different types of racks; each rack is uniquely identified with a 2D barcode. Figure 2 shows the eLabBench augmentation of a rack with a matrix-like visualization, and a user annotating a digital representation of a test tube on the augmented rack. Moreover, part of the experimental process involves the analysis of samples on machines biologists are not very familiar with,
Infrastructure – ABC Server & ActivityDock
The activity-based computing (ABC) Server infrastructure consists of a PHP application framework and API providing basic operations for the management of activities and resources: creation, modification, deletion, retrieval, storage, copy, annotation, and con- servation of state. The ABC server infrastructure represents activities and resources as XML documents, and communicates with the client applications through a REST protocol.
The activityDock is the client application allowing biologists to interact with their activities and resources from their personal computers. The dock is an Adobe Air application running on Windows, MacOSX and Linux. Activities and resources are synchronized transparently with the bench via the miniAbcLib communicating with the ABC server. The miniAbcLib also caches data so that the activityDock can be used when users are offline.
Researchers
The eLabBech was designed, implemented and evaluated together with Aurélien Tabard, Juan-David Hincapié-Ramos, Ebbe Slot Andersen, and Morten Esbensen.
References
![[pdf]](https://www.bardram.net/wp-content/plugins/papercite/img/pdf.png)
[Bibtex]
@inproceedings{its2011:tabard,
author = {Tabard, Aur{\'e}lien and Hincapi{\'e}-Ramos, Juan-David and Esbensen, Morten and Bardram, Jakob E.},
title = {The eLabBench: An Interactive Tabletop System for the Biology Laboratory},
booktitle = {Proceedings of the ACM International Conference on Interactive Tabletops and Surfaces},
series = {ITS '11},
year = {2011},
isbn = {978-1-4503-0871-7},
location = {Kobe, Japan},
pages = {202--211},
numpages = {10},
url = {http://doi.acm.org/10.1145/2076354.2076391},
doi = {10.1145/2076354.2076391},
acmid = {2076391},
publisher = {ACM},
address = {New York, NY, USA},
keywords = {activity-based computing, bench, biology, digital notebook, laboratory, tabletop},
}![[pdf]](https://www.bardram.net/wp-content/plugins/papercite/img/pdf.png)
[Bibtex]
@inproceedings{p3051-tabard,
author = {Tabard, Aurelien and Hincapie Ramos, Juan David and Bardram, Jakob E},
title = {The eLabBench in the wild: supporting exploration in a molecular biology lab},
booktitle = {Proceedings of the 2012 ACM annual conference on Human Factors in Computing Systems},
series = {CHI '12},
year = {2012},
isbn = {978-1-4503-1015-4},
location = {Austin, Texas, USA},
pages = {3051--3060},
numpages = {10},
url = {http://doi.acm.org/10.1145/2207676.2208718},
doi = {10.1145/2207676.2208718},
acmid = {2208718},
publisher = {ACM},
address = {New York, NY, USA},
keywords = {biology, design, digital bench, elabbench, experiment, field study, laboratory, life-sciences, research practices, tabletop},
tag={mini-grid,elabbench,conference},
pdf={p3051-tabard.pdf},
}![[pdf]](https://www.bardram.net/wp-content/plugins/papercite/img/pdf.png)
[Bibtex]
@inproceedings{p251-tabard,
author = {Tabard, Aurelien and Gurn, Simon and Butz, Andreas and Bardram, Jakob E.},
title = {A case study of object and occlusion management on the eLabBench, a mixed physical/digital tabletop},
booktitle = {Proceedings of the 2013 ACM international conference on Interactive tabletops and surfaces},
series = {ITS '13},
year = {2013},
isbn = {978-1-4503-2271-3},
location = {St. Andrews, Scotland, United Kingdom},
pages = {251--254},
numpages = {4},
url = {http://doi.acm.org/10.1145/2512349.2512794},
doi = {10.1145/2512349.2512794},
acmid = {2512794},
publisher = {ACM},
address = {New York, NY, USA},
keywords = {laboratory bench, object management, occlusion, tabletop, tangible interaction},
tag={mini-grid,elabbench,conference},
pdf={p251-tabard.pdf},
}
